-Search query
-Search result
Showing 1 - 50 of 186 items for (author: haynes & bf)

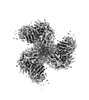
EMDB-40853: 
CH505 Disulfide Stapled SOSIP Bound to b12 Fab
Method: single particle / : Henderson R
EMDB-40854: 
CH505 Disulfide Stapled SOSIP Bound to CH235.12 Fab
Method: single particle / : Henderson R

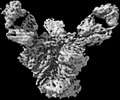
EMDB-27703: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P
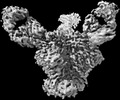
EMDB-27624: 
Cryo-EM structure of HIV-1 Env(CH848 10.17 DS.SOSIP_DT) in complex with DH1030.1 Fab
Method: single particle / : Gobeil S, Acharya P
EMDB-27628: 
Cryo-EM structure of HIV-1 Env(BG505.T332N SOSIP) in complex with DH1030.1 Fab
Method: single particle / : Gobeil S, Acharya P

EMDB-40273: 
CryoEM structure of VRC01-CH848.0358.80
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40274: 
CryoEM structure of VRC01-CH848.0836.10
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40275: 
CryoEM structure of DH270.6-CH848.0526.25
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40277: 
CryoEM structure of DH270.6-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40278: 
CryoEM structure of DH270.5-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40279: 
CryoEM structure of VRC01-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40280: 
CryoEM structure of DH270.4-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40281: 
CryoEM structure of VRC01-CH848.0526.25
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40282: 
CryoEM structure of DH270.UCA.G57R-CH848.10.17DT
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40283: 
CryoEM structure of DH270.UCA-CH848.10.17DT
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40284: 
CryoEM structure of DH270.3-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40285: 
CryoEM structure of DH270.I5.6-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40286: 
CryoEM structure of DH270.I4.6-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40287: 
CryoEM structure of DH270.I3-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40288: 
CryoEM structure of DH270.I2-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40289: 
CryoEM structure of DH270.2-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40290: 
CryoEM structure of DH270.1-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40291: 
CryoEM structure of DH270.I1.6-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P
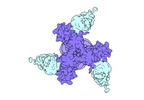
EMDB-27706: 
Vaccine elicited Antibody MU89 bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer
Method: single particle / : Stalls V, Acharya P
EMDB-27776: 
Vaccine elicited Antibody MU89+S27Y bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer
Method: single particle / : Stalls V, Acharya P

EMDB-28608: 
Cryo-EM structure of CH848 10.17DT DS-SOSIP-2P Env
Method: single particle / : Wrapp D, Acharya P, Haynes BF

EMDB-24065: 
CryoEM map of monoclonal antibody Fab DH1025.1 bound to bound to CH505.M5 SOSIP trimer
Method: single particle / : Manne K, Acharya P
EMDB-26676: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-26677: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains)
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-26678: 
An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-27043: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody SP1-77 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K
EMDB-27044: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-7-53 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K

EMDB-27046: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-5-82 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K

EMDB-26961: 
Triple mutant (K417N-E484K-N501Y) SARS-CoV-2 spike protein in the 3-RBD-Down conformation (S-GSAS-D614G-K417N-E484K-N501Y)
Method: single particle / : Gobeil S, Acharya P

EMDB-26433: 
SARS-CoV-2 Omicron-BA.2 3-RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P
EMDB-26435: 
SARS-CoV-2 Omicron-BA.2 3-RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P

EMDB-26436: 
SARS-CoV-2 Omicron-BA.2 3-RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P

EMDB-26643: 
SARS-CoV-2 Omicron-BA.2 1.5-RBD up Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P

EMDB-26644: 
SARS-CoV-2 Omicron-BA.2 1-RBD-up Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P

EMDB-26647: 
SARS-CoV-2 Omicron-BA.2 1-RBD-up Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P

EMDB-26600: 
SARS-CoV-2 Omicron-BA.1 2-RBD up Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.1)
Method: single particle / : Stalls V, Acharya P

EMDB-25880: 
SARS-CoV-2 Omicron 1-RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron)
Method: single particle / : Stalls V, Acharya P

EMDB-25983: 
SARS-CoV-2 Omicron 3-RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron)
Method: single particle / : Stalls V, Acharya P

EMDB-25984: 
SARS-CoV-2 Omicron 1-RBD up Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron)
Method: single particle / : Stalls V, Acharya P

EMDB-25846: 
SARS-CoV-2 Omicron 1-RBD up Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron)
Method: single particle / : Stalls V, Acharya P

EMDB-25865: 
SARS-CoV-2 Omicron 3-RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron)
Method: single particle / : Stalls V, Acharya P

EMDB-25904: 
CryoEM map of SARS-CoV-2 S protein in complex with Receptor Binding Domain antibody DH1042
Method: single particle / : Manne K, May A, Acharya P

EMDB-25893: 
Structure of RBD directed antibody DH1042 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interface
Method: single particle / : May AJ, Manne K, Acharya P

EMDB-26038: 
Delta (B.1.617.2) SARS-CoV-2 variant spike protein (S-GSAS-Delta) in the 3-RBD-down conformation; consensus state D1
Method: single particle / : Gobeil S, Acharya P

EMDB-26039: 
Delta (B.1.617.2) SARS-CoV-2 variant spike protein (S-GSAS-Delta) in the 1-RBD-up conformation; consensus state D2
Method: single particle / : Gobeil S, Acharya P
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
